Technical reply to the criticism of Dr. Bucci

We report below the technical response of Dr. Loretta Bolgan to the criticism published by Dr. Bucci on Corvelva analyzes.
Dear Dr. Bucci,
below I respond to the critical issues you raised in particular in your article published on: BadScientists.com
First of all I would like to point out to you that the use of NGS / HTS (Next Generation Sequencing or High-Throughput Sequencing) technologies is NOT original on complex substrates of biotechnological origin, such as vaccines.
NGS technology has already been used in vaccine products, for example:
- in the analysis of commercial batches of the rotavirus vaccine (Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, et al. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol 2010; 84: 6033–40) allowing the subsequent blocking of the marketing of lots contaminated with Porcine Circovirus type 1 (PCV1), despite previously having passed all the tests required for development, clinical trials and production (Dubin, G .; Toussaint, JF; Cassart, JP; Howe, B .; Boyce, D .; Friedland, L .; Abu-Elyazeed, R .; Poncelet, S .; Han, HH; Debrus, S. Investigation of a regulatory agency inquiry into potential porcine circovirus type 1 contamination of the human rotavirus vaccine, Rotarix: approach and outcome. Hum. Vaccines Immunother. 2013, 9, 2398–2408).
- in the detection of a new Rhabdovirus in the Sf9 cell line (MH, Galvin TA, Glasner DR, Shaheduzzaman S, Khan AS. Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines. J Virol 2014; 88: 6576–85)
- in identifying the mumps vaccine virus in the brain of an immunocompromised child who died of encephalitis (Morfopoulou S, Mee ET, Connaughton SM, Brown JR, Gilmour K, Chong WK, Duprex WP, Ferguson D, Hubank M, Hutchinson C, Kaliakatsos M, McQuaid S, Paine S, Plagnol V, Ruis C, Virasami A, Zhan H, Jacques TS, Schepelmann S, Qasim W, Breuer J. Deep sequencing reveals persistence of cell-associated mumps vaccine virus in chronic encephalitis. Acta Neuropathol. 2017 Jan; 133 (1): 139-147. Doi: 10.1007 / s00401- 016-1629-y. Epub 2016 Oct 21. PubMed PMID: 27770235).
Also in the 2017 collaborative work 'A Multicenter Study To Evaluate the Performance of High-Throughput Sequencing for Virus Detection' (A Khan AS, Ng SHS, Vandeputte O, Aljanahi A, Deyati A, Cassart JP, Charlebois RL, Taliaferro LP. A Multicenter Study To Evaluate the Performance of High-Throughput Sequencing for Virus Detection. MSphere. 2017 Sep 13; 2 (5) .pii: e00307-17. Doi: 10.1128 / mSphere.00307-17. ECollection 2017 Sep-Oct. PubMed PMID : 28932815; PubMed Central PMCID: PMC5597969), is highlighted as analyzes performed with different HTS instruments, with different protocols of sample preparation and data analysis, on samples mimicking complex biological materials, have shown a similar sensitivity in the detection of viruses in 3 several workshops.
Finally, in the 'Report of the international conference on next generation sequencing for adventitious virus detection in biologicals' (Khan AS, Benetti L, Blumel J, Deforce D, Egan WM, Knezevic I, Krause PR, Mallet L, Mayet D, Minor PD , Neels P, Wang G. Report of the international conference on next generation sequencing for adventitious virus detection in biologicals. Biologicals. 2018 Sep; 55: 1-16. Doi: 10.1016 / j.biologicals.2018.08.002. Epub 2018 Aug 6 PubMed PMID: 30093175), report of an international conference held in Rockville (MD) from 26 to 27 October 2017, co-organized by IABS (International Alliance for Biological Standardization) and the FDA (US Food and Drug Administration), in attended by one hundred and twenty-eight scientists from 16 different countries, including researchers Glaxo SmithKline, Merck and Sanofi, it is stated that:
“The main advantages of the NGS for safety tests for virus detection are the rapid and non-targeted approach, which can be used in multiple substrates and which allows to detect a wide range of viruses, including variants and new species. ; and that there is no need for specific amplification, while PCR requires targeting a specific sequence "
And yet:
"HTS technologies are revealing some of the current limitations of programs to test biological materials and could integrate gaps in these programs and increase product safety"
And in the conclusions of the report we read:
"Continuous collaborative and scientific efforts, exchanges between regulatory agencies and other public bodies, industry, academic laboratories and service providers will advance the field of NGS with the aim of guaranteeing the safety of organic products that impact on human and animal health"
I reply below to your observations, in particular to those that allow us to better explain the preliminary data that we have produced on some lots of Priorix Tetra.
Remark 1
"One wonders how this practice of secrecy, contrary to any criterion of data sharing, can be reconciled with the continuous requests for transparency made by the CORVELVA association and by the president of the National Order of Biologists in terms of vaccine safety: it is lawful launch an alarm, without providing all the data necessary to evaluate its consistency?
How can the scientific community and citizens assess the weight of certain claims, if the complete original data is kept secret, pending a public prosecutor to decide whether and how to proceed with a judicial action? "
Answer 1
The metagenomics data disclosed by Corvelva for the Priorix Tetra (MPRV vaccine) and discussed in the report published in December were preliminary (an updated version was disclosed on the Corvelva website on January 23, the result of a further step in data analysis), because, as already reiterated on several occasions, the data disclosed so far result from a process of research and technological development started in 2017, with the aim of fine-tuning and optimizing the whole procedure, from the processing of the sample to the bioinformatics analysis. It was decided to divulge the results during the work, as they become available, in absolute transparency (just the opposite of what is declared by you as a 'practice of secrecy'!) And in any case strongly supported by the scientific literature on the 'topic.
In our opinion, what we have disclosed so far about this vaccine, especially regarding the high amount of human fetal DNA, is nothing new or unexpected and least of all unknown to pharmaceutical companies.
Remark 2
“It can easily be seen that the total of reads indicated at the top left (highlighted in yellow) is over 6 million; however, the sum of all the reads divided by class of organism is approximately 5 and a half million, with a "shortfall" of over 700.000 reads (and a percentage sum, indicated by the red box, equal to 88%). This simple addition, repeated on all tables, asks the question of where the missing reads ended up (a percentage that is certainly not negligible) and why they have not been reported. The possibility that the researchers made a mistake remains clearly open "
Answer 2
Let's say that 700.000 reads more or less slightly change the substance of the preliminary screening analyzes carried out. Missing sequences are included in the calculation of 'unassigned' sequences. As already mentioned, the data are not definitive and there may be inaccuracies in the reports. Everything will be reviewed and corrected as the work progresses.
Remark 3
“The rubella virus genome, one of the attenuated viruses needed to confer immunogenicity, would not be present. The detection at very low levels, bordering on statistical noise, of a microorganism in the sample under examination does not therefore depend on its absence, but on the non-optimized identification method. The same, identical difficulty can be encountered for the identification of any other genome, for which the method of analysis has not been suitably calibrated; the lack of disclosure of the rubella virus genome (moreover an encapsulated single-helix virus) is therefore attributable to the method used, without the need to invoke its absence in the vaccine lots examined "
Answer 3
His thesis is acceptable. We are in the research and development phase, however the technology is very robust and recognized by the scientific community that deals with deep sequencing for the detection of viruses in complex biological substrates. Inter-laboratory tests and the introduction of appropriate certified viral mix standards will allow us to verify or disprove the hypothesis you proposed, that is, that the method is not (yet) optimized for viruses.
In any case, the rubella genome was then found in the batch under examination, increasing the sequencing depth (114 paired-end sequences 125bp long, out of about 260 million sequences produced) as described in the second report of January 2019. Also in the vaccine MMRVax Pro from Merck, analyzed in 2017 with the same method, the rubella virus genome was found without having to go to such deep depths, proving that the method works.
Remark 4
“The genomes of numerous contaminating organisms belonging to numerous taxa would be detected, including helminths, bacteria and adventitious human pathogenic viruses. There is also a further problem in the tables presented, consisting of the abundant frequency of sequences attributable to viruses integrated in the human genome (endoviruses of various nature) or retroviral sequences known to be found in the normal human genome (this is the case, for example, of sequences integrated retrovirals that can be mistaken for HIV) or in the genome of chicken embryonic cells used for vaccine production, typically found as false assignments to contaminating genomes in DNA or RNA sequencing depending on the particular endosequence considered. First of all, there are numerous species listed in the report tables, which are found on the basis of a number of reads fully included in the statistical noise. 3 or less reads is a risky procedure that leads to a high number of false positives "
Answer 4
The version of the report disclosed on the Corvelva website in December, to which you refer, has been updated recently with a version in which a part of the contaminants was preliminarily validated with alternative software, and then manually. The first analysis had been deliberately performed without filters to highlight every possible signal and all the problems related to an 'open' analysis. The purpose of our work is not, as reiterated several times, to carry out an analysis of the batch release, but to carry out a preliminary screening, and subsequently an inter-laboratory confirmation on the critical issues that have emerged, through the use of a well-established technology in the field. genomic, already applied on vaccines and forthcoming also by regulatory agencies and large pharmaceutical companies to increase the quality and consequently the safety of the product.
If you define the limit of 3 reads (based on what?), Then we could at least think the signs of presence of endogenous retroviruses in this vaccine are likely (Human Endogeneous Retrovirus K: 32 reads of 125bp, corresponding to 4000bp and HERVH- env 62: 4 reads, corresponding to 500bp), in the RNA-seq data, which by definition represent the transcribed and therefore potentially active material.
Instead, I agree with you that the retroviral sequences found in the DNA-seq data can be proviruses integrated into the human genome, given the very high amount of human DNA in the vaccine in question.
I remember that in the Attenuvax vaccine (measles vaccine), 4 sequences covering 700bp (Victoria JG, Wang C, Jones MS, Jaing C, McLoughlin K, Gardner S, et al. Viral nucleic acids in live-attenuated vaccines: detection of minority variants and an adventitious virus. J Virol 2010; 84: 6033–40) allowed to detect by NGS an Avian Leukosis Virus (ALV) virus contamination for the first time in a vaccine, then confirmed by nested PCR.
Remark 5
"As regards the first of these potential contaminants, the non-pathogenic nitrogen fixing bacterium Bradyrhizobium, its" appearance "in sequencing laboratories it is well known as a problem related to the preparation of the samples to be sequenced, attributable to contamination of DNA purification kits, water and reagents to be used. A well-known problem, in which in the work of "exploratory" genomics a control sample, for example containing water, is sequenced in parallel to the sample under examination "
Answer 5
The whites were made (both from extraction and from the library), but libraries were not obtained and therefore it was not possible to run them on the sequencer to check exactly what the reagent / environmental contamination is. However efforts are being made to try to understand what 'physiological' contaminations are and inter-laboratory tests will also help in determining possible laboratory-dependent bacterial contaminations.
Remark 6
“As for the two types of worms revealed in RNA sequencing analysis, it is sufficient to observe how they are not present in the corresponding DNA sequencing analyzes, where they should be much better detectable; consequently, the actual presence of the two species appears as an artifact, which cannot be confirmed in the light of the same DNA sequencing data presented.
In light of the considerations made, it is therefore not possible to confirm the presence of any of the contaminating genomes reported, and there are strong clues that lead us to think about the presence of numerous false positives "
Answer 6
The second version of the report has already provided for a first validation of some organisms found, both with alternative software, and manually.
The evaluation of the incorrect attributions of the Kraken software (moreover one of the most commonly used software by the scientific community for whole-genome metagenomic analyzes) is being used to develop an ad hoc bioinformatics pipeline for this type of analysis, which does not require the use of others. software and manual inspection (moreover common in the field of metagenomics for state of the art problems due to the absence of specific and particularly accurate databases).
Remark 7
"The human DNA detected would be of high molecular weight and the coverage of the human genome would be total, so that the entire genome of human fetal cells and not portions of it would be present in the vaccine lots examined.
"In the Priorix Tetra vaccine the human genomic DNA is high molecular weight (> 10.000bp) and the total sequential coverage of the entire reference human genome (HG-19) shows that it is the entire genome of the fetal cells used to the vaccine virus culture to be present and not just portions of it. "
No direct evidence is given for the stated statement, without prejudice to the presence of DNA weighing around 100.000 bases revealed by gel electrophoresis by the authors (documented with a poor quality figure in the CORVELVA document, of which it would be advisable to obtain a high original resolution to assess whether the data is presented in an intact manner).
However, the authors forget that the DNA of some of the attenuated viruses present in the vaccine in question is about the same size as what they revealed in the gel (for example, the DNA of the chickenpox virus is 125 kb); therefore, even if you want to glimpse traces of high weight DNA in the gel image presented, this can clearly be attributed to the DNA of the attenuated viruses present in the vaccine, and not to fragments of human DNA. Again, we are faced with an overinterpretation of the experimental result. As for the second element on which the CORVELVA claim is based, that is that the entire human genome would be present in vaccines, we remember that there is not even need to have access to the detailed coverage data to weigh its consistency: depth of sequencing used and with the number of reads obtained, this statement is totally implausible. Furthermore, the sequencing technique used, based on short sequences, 125 nucleotides, does not allow to define if a genome is fragmented. The sequencing technique used only allows to estimate the fraction of sequenced genomic DNA "
Answer 7
The human DNA in this vaccine is approximately 8 to 1 relative to the chickenpox DNA (88% of the reads are of human origin, compared to 11% of the chicken pox in the last batch analyzed A71CB256A). Other attenuated dsDNA viruses are not in the vaccine because measles, mumps and rubella viruses are single-stranded RNA viruses that cannot be viewed on agarose gel. NGS is a quantitative technology, therefore a simple fluorimetric quantification of the total DNA extracted from the vaccine (e.g. lot. A71CB256A = 3,7 micrograms per dose), associated with the consideration of relative quantification made above (8: 1), allows us to be able to say that cellular DNA is about 2,9 micrograms per dose, compared to about 0.74 micrograms of chickenpox DNA. It is therefore plausible that at least a portion of the high molecular weight DNA seen on gel may be that of the cells of the fetal cell line MRC5.
The direct evidence that within this product there is a COMPLETE human genome (i.e. with non-coding genes and sequences), fragmented or not, is given by the result of the alignment of the human-derived reads on the human reference hg19.
The following table, highlighted in orange, shows the result expressed in 'Av_cov = average coverage' of the alignment of the human sequences of the 3 batches of Priorix tested (1st, 2nd and 3rd lot) on human chromosomes. In column 1, chrM is mitochondrial DNA, while Chr1 to ChrY are the assembled human chromosomes, including the sex chromosomes X and Y. Column 2 shows the length of the assembled human chromosomes expressed in base pairs.
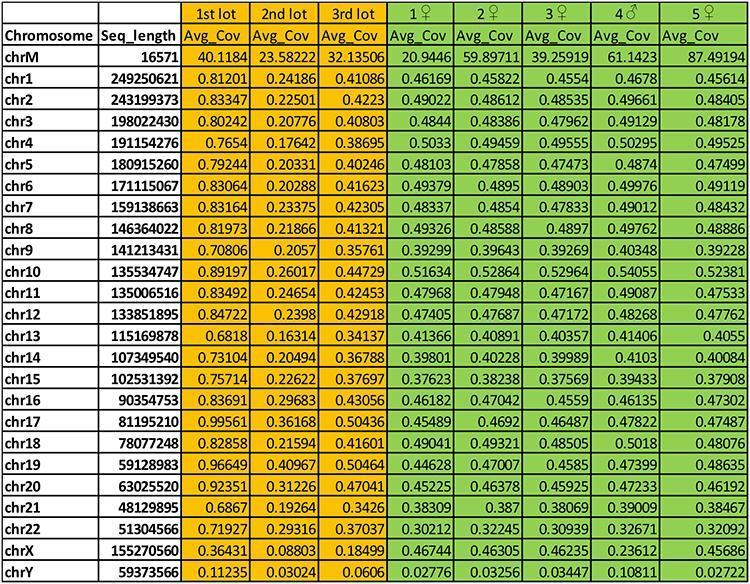
The coverage is low (Avg_cov on average along each chromosome <than 1x) but the homogeneity of distribution of the reads that align uniquely, along all human chromosomes and the presence also of reads that align with higher coverage on the genome mitochondrial (in human cells there are 2 genomes, one nuclear about 3Gbp large and divided into chromosomes and one smaller, circular, about 16Kbp mitochondrial), however, allows us to indisputably recognize a situation similar to a low-pass genome sequencing of a human genome individual. For an easier understanding of what is stated, in the part indicated in green there are some human low pass whole genome sequencing (5 samples, 4 females and 1 male) at a depth similar to that produced for the 3 vaccine lots of Priorix Tetra.
An indication of the sex of the individual (most likely male) in the vaccine can also be deduced from the relationship between the average coverage for the X and Y chromosome.
The human DNA contained in the previously sequenced lots of Priorix has also been qualified as belonging to the MRC-5 fetal line, that is the continuous cell line from lung tissue of a male abortive fetus of the 60s, in which the virus of the chickenpox and rubella. The analysis of the variants of the mitochondrial DNA present in the vaccine compared to the mitochondrial DNA of the MRC-5 line (the DNA of the cell line was purchased from ATCC, the main resource of standard biological materials in the world) shows that they are the same individual.
Remark 8
"There has long been evidence that even very high doses of human DNA cannot induce significant risks even over long observation periods, as confirmed by studies on non-human primates, from clinical experience with vaccines and other biological drugs and from recent risk assessment studies which suggest that even in those non-European countries where there are thresholds for the amount of injectable DNA, these thresholds are unnecessarily stringent "
Answer 8
The 1995 article (https://www.sciencedirect.com/science/article/pii/S104510568570036X?via%3Dihub) cited by you, concerns a study on primates, which in my opinion is certainly not sufficient to demonstrate the absence of infectivity, oncogenicity and autoimmunity of human DNA injected into children, as stated by the same authors in the final part of the discussion. In the experiment, a large amount of human DNA is injected into monkeys, therefore not DNA of the same species.
The second cited article (https://www.ncbi.nlm.nih.gov/pubmed/23569076) states very clearly that the residual DNA is infectious at 2 micrograms, even if in any case this limit is obtained from a statistical evaluation and not from experimental data. In any case, following the author's indications, the Priorix vaccine would have a quantity of fetal DNA clearly in line with that determined as at risk of infectious disease.
Merck for the Varivax vaccine (chicken pox, live attenuated virus) declares in the American leaflet the presence of human fetal DNA derived from MRC-5 cells which, from the analyzes carried out by Dr. Deisher, appears to be in an amount of about 2 micrograms.
In the case of the Italian leaflet of the Priorix Tetra, however, the presence of DNA of the MRC-5 line is not indicated, despite being in quantities of the same order of magnitude as that contained in the American Varivax. In any case 2 micrograms of fetal DNA of MRC-5 is not in my opinion a residual quantity, but a real component of the vaccine.
The in vitro studies of Dr. Deisher, which we hope will be published shortly, unfortunately show puzzling results when a vaccine containing fetal human DNA (hypomethylated) is incubated in vitro with human hematopoietic stem cells. But we wait for the research to be published and then we can meet again on the topic.
Loretta Bolgan *
* Doctor in chemistry and pharmaceutical technologies, with a doctorate in pharmaceutical sciences from Harvard medical school Boston. She worked in the pharmaceutical industry sector where she dealt with the registration and development of research projects in the oncology field. Consultant part of law 210/92, environmental pollution and occupational diseases, participated in the last parliamentary commission of inquiry on depleted uranium in the vaccine group. Current consultant for the National Order of Biologists for the toxicology of drugs and vaccines, he also deals with nutrition and complementary therapies.
IMPORTANT NOTE: Corvelva invites you to get in-depth information by reading all the sections and links, as well as the manufacturer's product leaflets and technical data sheets, and to speak with one or more trusted professionals before deciding to vaccinate yourself or your child. This information is for informational purposes only and is not intended as medical advice.

