Short presentation of the results
Today we present a further and very important in-depth study on the genomic sequencing of the cell line we found within the quadrivalent MPRV vaccine (measles, rubella, mumps and chickenpox) Priorix Tetra®, and that of the MRC-5 cell line (the one declaredly used in the development of the vaccine itself).
Summarizing previous published reports:
- First, we demonstrated that the amount of DNA present in the above-mentioned vaccine is well above the allowed threshold;
- Secondly,we verified that the cell line is actually the declared MRC-5; 2
- Lastly, we carried out the sequencing of the cell line genome directly extracted from the vaccine, highlighting major mutations that open to the possibility of serious health risks in using this vaccine.
What we are presenting here is precisely a comparative work between the two genomes: the one we actually found inside the vaccine and the one purchased from the international cell line bank as MRC-5.
The result is that the two lines display important differences from the point of view of genetic stability. In particular, the vaccine genome is significantly modified compared to the cell line registered in 1966.
That means the producers, in this case of Priorix Tetra®, have purchased the cell line under optimal conditions from the point of view of genetic stability and, by the over-time continuous use for production purposes, have marketed vaccines containing progressively more and more modified human genetic material, that is dangerous for the health of the vaccinees themselves.
Since the regulatory agencies do not require periodic monitoring of the genetic stability of foetal lines and do not even set a minimum safety threshold, we can deduce that a fundamental requirement for the quality and safety of vaccines obtained from human cell lines has not been met for some time now, namely the need to verify that they do not develop potentially dangerous mutations and rearrangements and to ensure the elimination of processing genetic material residues, if present.
It is not our responsibility to identify which part of the production or preservation process could affect the product quality and safety, but what we can confirm today, by publishing this report, is the urgency to carry out an in-depth examination of our analyses results we have been notifying in the last two years, focusing on the finished product, sold and administered and not only on the single authorized "ingredients".
We would like to remind once again that these products are part of the Italian vaccination schedule and are currently mandated on the paediatric population in our country as well as in other European countries and in general throughout the world.
The regulatory agencies have a huge responsibility with regard to safety, which is currently being put into words but, in practice, and as far as our analyses show, cannot be guaranteed and is in fact clearly not guaranteed.
Circular display of genomes (circos plot)
A graphical representation of the two genomes called 'circos plot' is shown below.
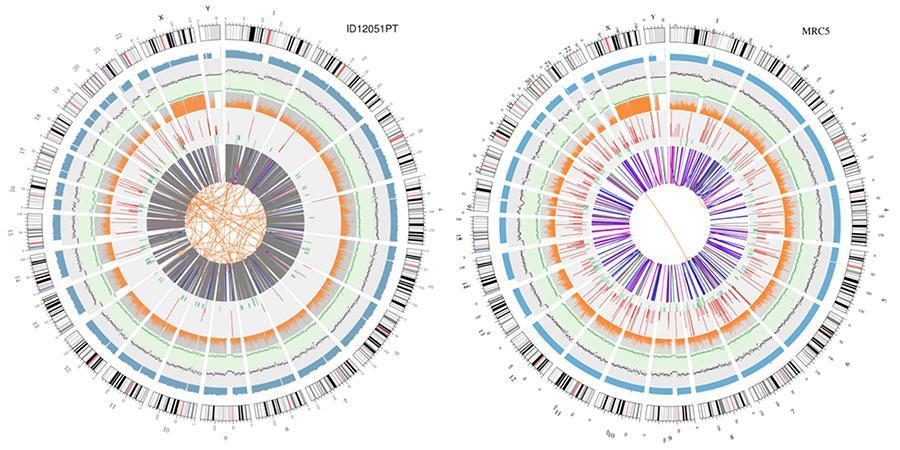
Explanation of the various concentric circles
- The outer circle (the first circle) is the chromosome number.
- The second ring (blue) represents the coverage of histogram style reads. Each histogram is the average coverage of an area of 0.5 Mbp.
- The third ring (black) represents the density of the INDELs in 'dispersion graph' style. Each black dot is calculated as the number of INDELs (small insertions/deletions) in a range of 1 Mbp.
- The fourth ring (green) represents the snp density in 'dispersion graph' style. Each green dot is calculated as the number of SNPs (single nucleotide variants) in a range of 1 Mbp.
- The fifth ring represents the proportion of SNPs in homozygosity (orange) and heterozygosity (grey) histogram style. Each histogram is calculated from a 1 Mbp region.
- The sixth ring represents the inference of CNV (Variants in number of copies). Red means gain of DNA pieces and green means loss.
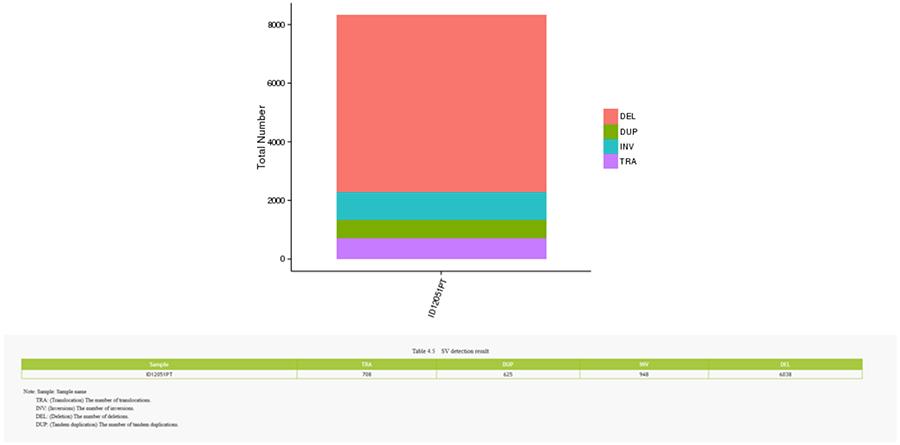
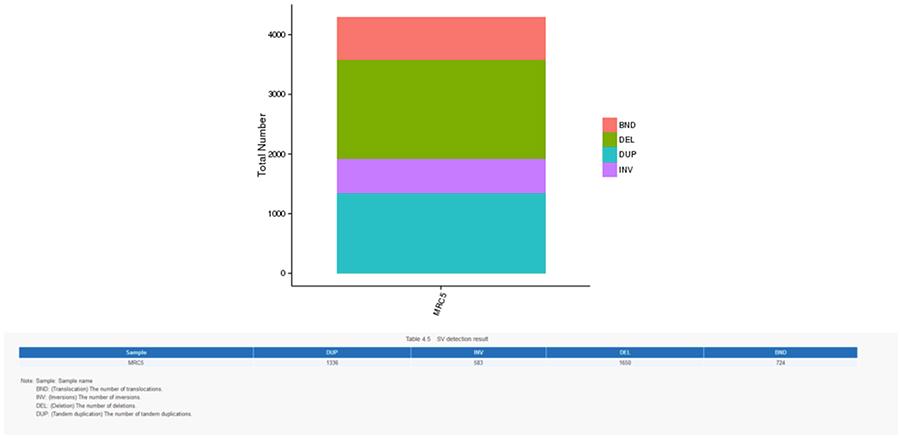
- The most central ring represents the inference of SV (Structural Variants) in the hexagonal and splicing areas. TRA (orange, translocations), INS (green, insertions), DEL (deletions, grey), DUP (duplications, pink) and INV (inversions, blue).
Conclusion
The two genomes display important differences. In particular, the vaccine genome appears to be seriously mutated compared to the cell line deposited in 1966. It must be said that we have not had the possibility to sequence the cell line used by GlaxoSmithKline for the production of Priorix Tetra® vaccine, but the one of the original stock.
Pharmaceutical companies using cell lines for the production of drugs and vaccines have their own MRC-5 line, but this fact does not change our doubt, that is, there is no periodic request for genetic stability monitoring, and this is a major issue in our view, especially because this is allowed without a threshold limit.
Download: CORVELVA-Priorix-Tetra-human-genome-and-MRC-5-cell-line.pdf
Priorix Tetra® human genome and MRC-5 cell line – comparative study (ATCC standard MRC-5 ATCC® CCL-171™)
The presence of rubella virus was demonstrated by sequencing an RNA-seq library with extremely high depth (approximately 260 million sequences Illumina produced). 114 sequences of 260 million have been detected, equal to 0.00004% of the total sequences. The rubella genome sequences have then been confirmed manually with the BLAST software (Basic Local Alignment Search Tool, https://blast.ncbi.nlm.nih.gov/Blast.cgi). The library sequenced at lower depth (about 12 million of match sequences Illumina, equal to 6 million fragments of library sequenced) had not detected the presence of ANY reads attributable to rubella, in this batch.
Other virus sequences present in the vaccine (chickenpox, measles and mumps) have also been validated by the same manual way, confirming they were correctly assigned.
Introduction
The new generation sequencers have become tools of choice for detailed analysis in the field of biology and medicine, especially the precision one. These tools allow a new and more global approach to a series of applications such as de novo sequencing, metagenomics, epigenomics, transcriptome sequencing and genome re-sequencing.
The latter application (re-sequencing) is widespread in the human field both for research and diagnostic purposes and consists in the sequencing with NGS (Next Generation Sequencing) technology of an entire individual genome in order to map single nucleotide mutations (SNP, pronunciation 'snip '), insertions and deletions of more or less long sequences occurred in certain positions of the genome and variations in the number of copies of portions of genome / genes (CNV, Copy Number Variants).
The sequencing procedure requires that the DNA of an individual is mechanically broken into fragments of small size (400-500 bp) and these fragments are linked to tracts of artificial DNA called adapters, which allow to fix the fragments of human DNA to a glass surface on which the bases are then read (A, C, G, T).
The DNA reading is obtained by means of incorporating chemical reactions of nucleotides labelled with fluorescent molecules. The millions of sequences (reads) that are obtained by sequencing occurred on the glass surface, are then mapped to the human genome reference with appropriate software, and then all the variants present in the analysed genome compared to the reference are identified.
This same procedure was performed on the human genome present in Priorix Tetra® batch n. A71CB256A and on the DNA extracted from the MRC-5 cell line, registered in 1966 at ATCC (MRC-5 ATCC® CCL-171 ™).
ATCC is the leading worldwide organization of resources and standards of biological materials whose mission focuses on the acquisition, authentication, production, preservation, development and distribution of reference microorganisms, cell lines and other biological standard materials. It was established in 1925, when a committee of scientists recognized the need for a centralized collection of biological materials that could serve scientists around the world.
The MRC-5 cell line is derived from normal lung tissue from a 14-week-old male foetus and was deposited in ATCC by J.P. Jacobs in September 1966. These cells are declared to be able to double from 42 to 46 populations before the senescence starts. The reference article for this cell line dates back to 1970: https://www.nature.com/articles/227168a0
Results
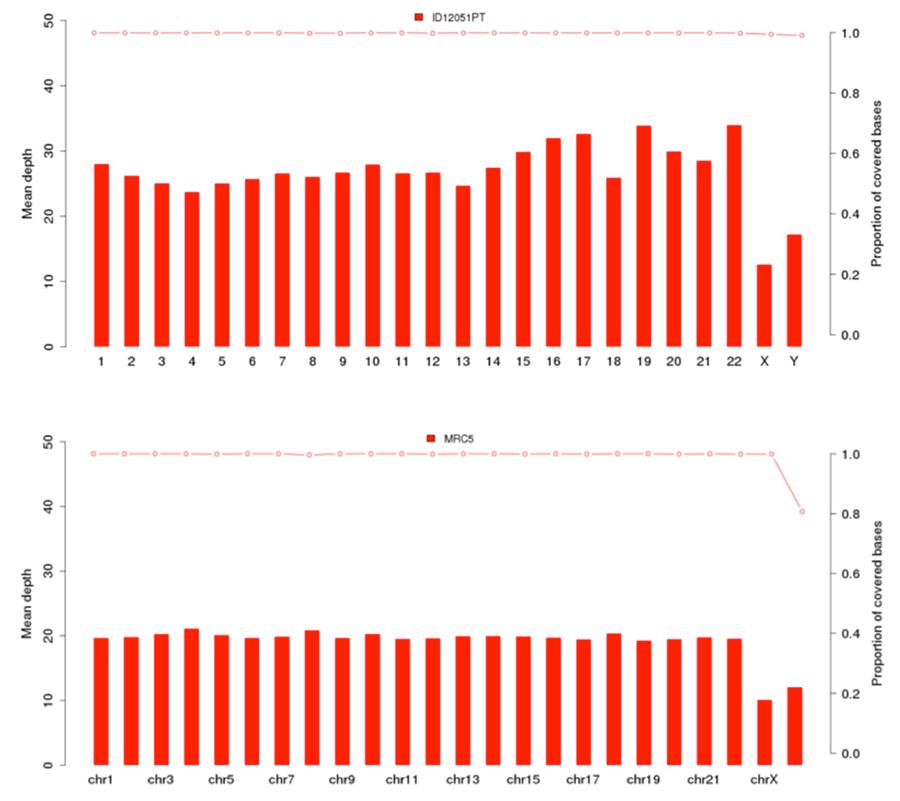
As previously shown, the human DNA present in Priorix Tetra® lot. n. A71CB256A (indicated in the following graphs as “sample ID12051PT”) is a complete individual genome, that is to say that the genomic DNA of all the chromosomes of a male individual is present. The coverage graphs (Mean depth = average coverage) of the 22 human chromosomes and of the X and Y chromosomes are shown below, for the vaccine (image above) and the MRC5 cell line (image below).
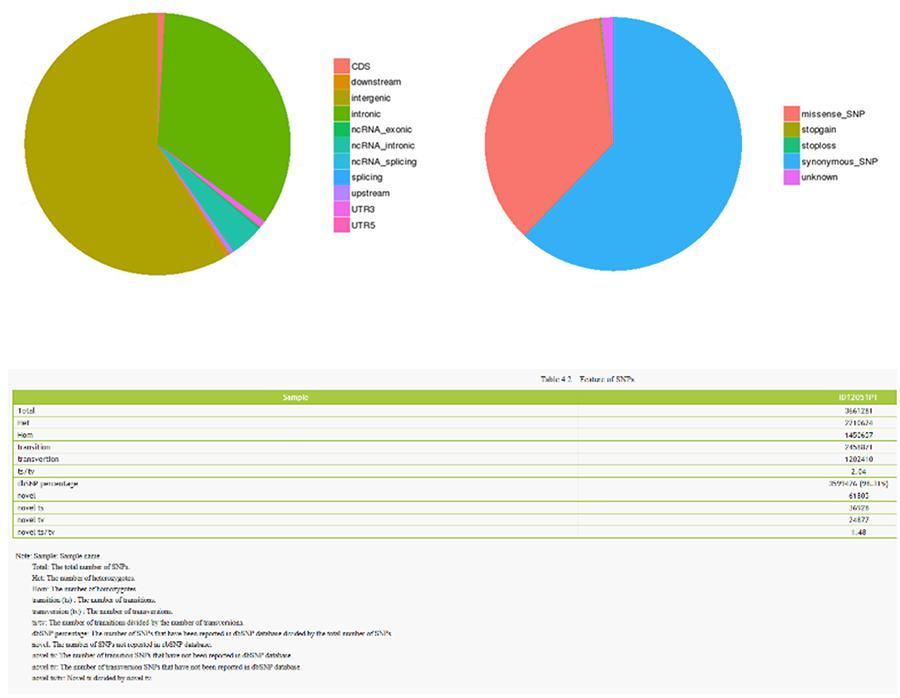
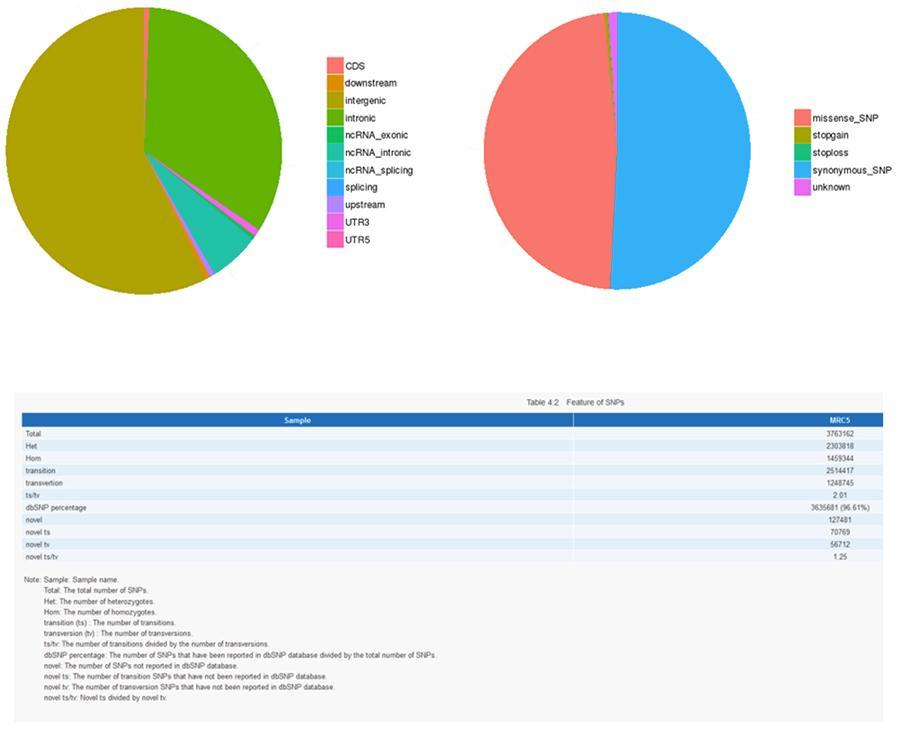
Below are shown the results of the analysis of the various types of variants.
Single nucleotide polymorphism (SNP) and short insertions / deletions (InDels)
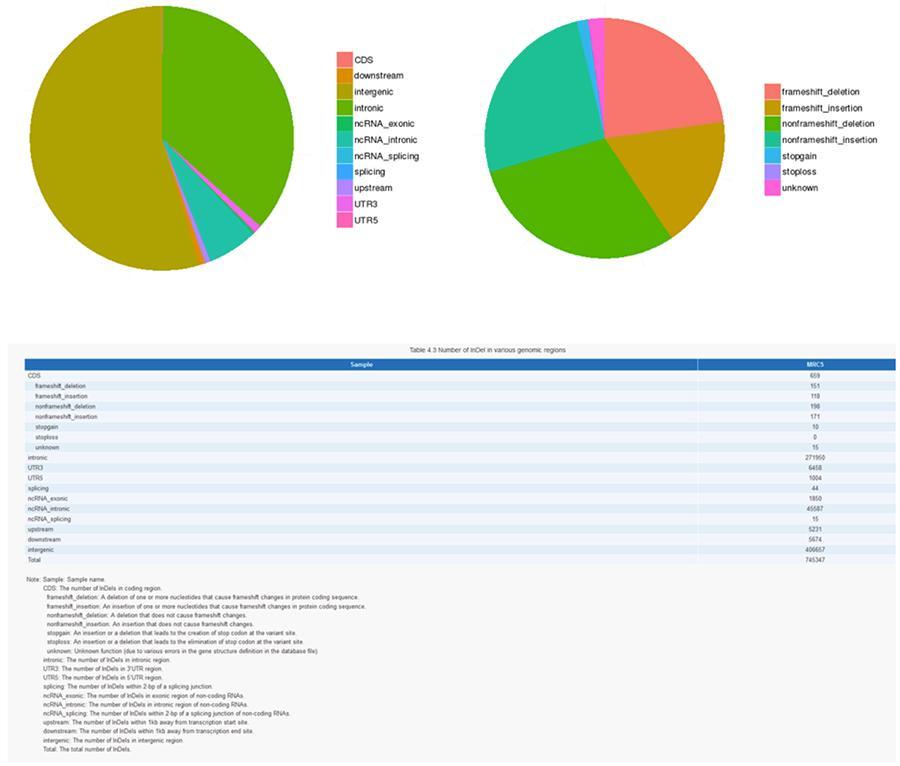
The variants of single individual DNA (SNP, pronounced 'snip') are polymorphisms, that is to say variations of the genetic material, borne by a single nucleotide. The 'InDels' are small insertions and deletions of less than 50bp in length and constitute another class of genomic variants in the human genome.
SNP (single nucleotide polymorphism) - ID12051PT
SNP (single nucleotide polymorphism) - MRC-5
InDels (short insertions / deletions) - ID12051PT
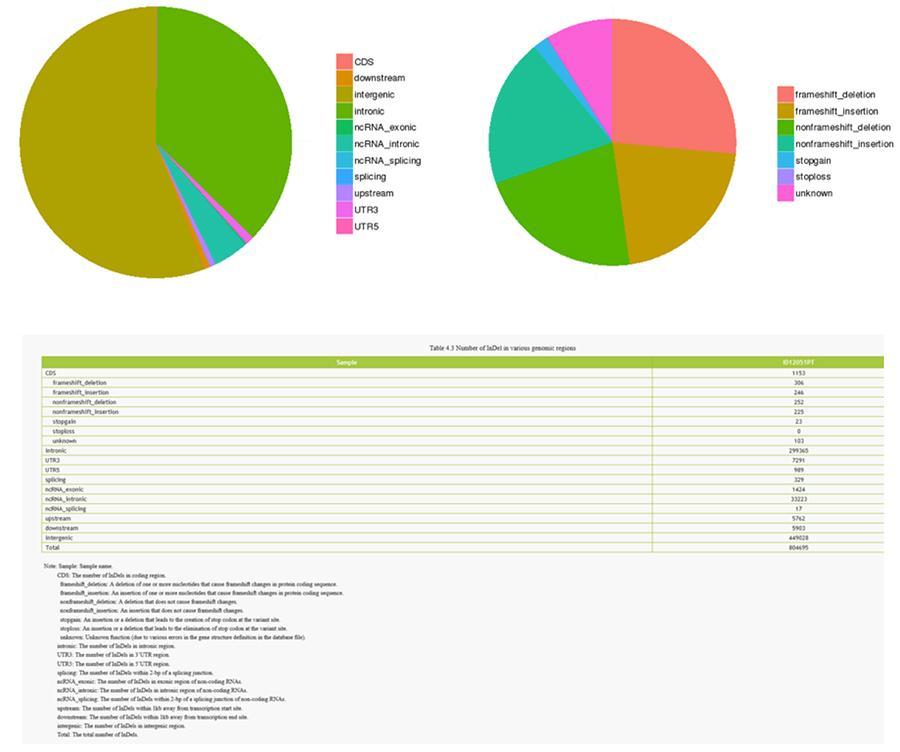
InDels (short insertions / deletions) - MRC-5
CNV (Copy Number Variants) and SV (Structural Variants)
Copy number variants (CNV) are genomic variants due to variations in the number of copies of relatively large fragments (longer than 50 bp) in individual genomes. There are two types of CNV: 'gain' type (copies gain) and ‘loss’ type (copies loss).
Structural variants (SV) are genomic variants with relatively large dimensions (>50 bp) including deletions, duplications, insertions, invertions and translocations.
CNV (copy number variants)
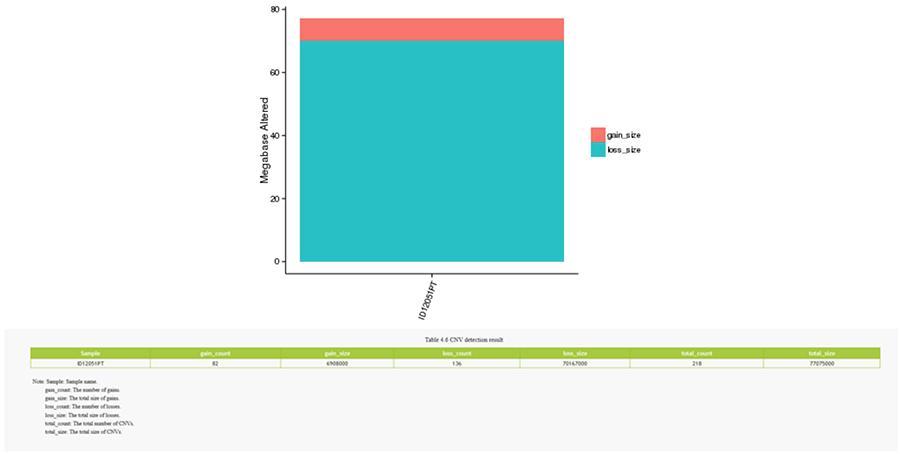
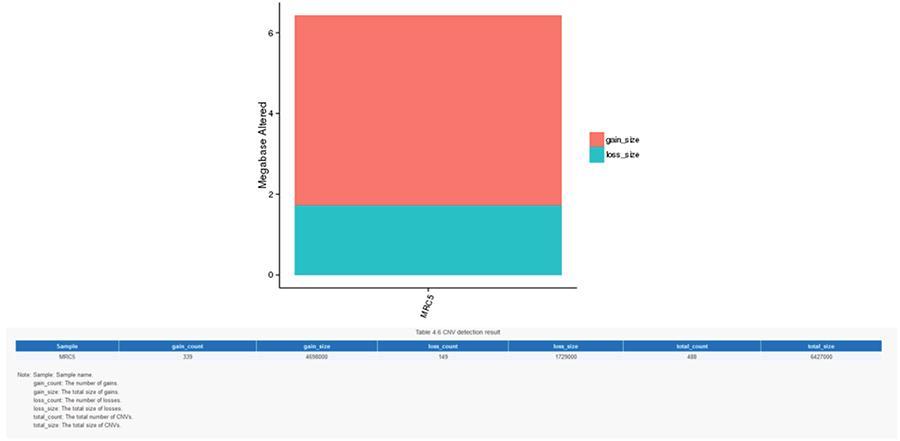
SV (structural variants)
Circular visualization of genomes (circos plot)
A graphical representation of the two genomes called (circos plot) is shown below.
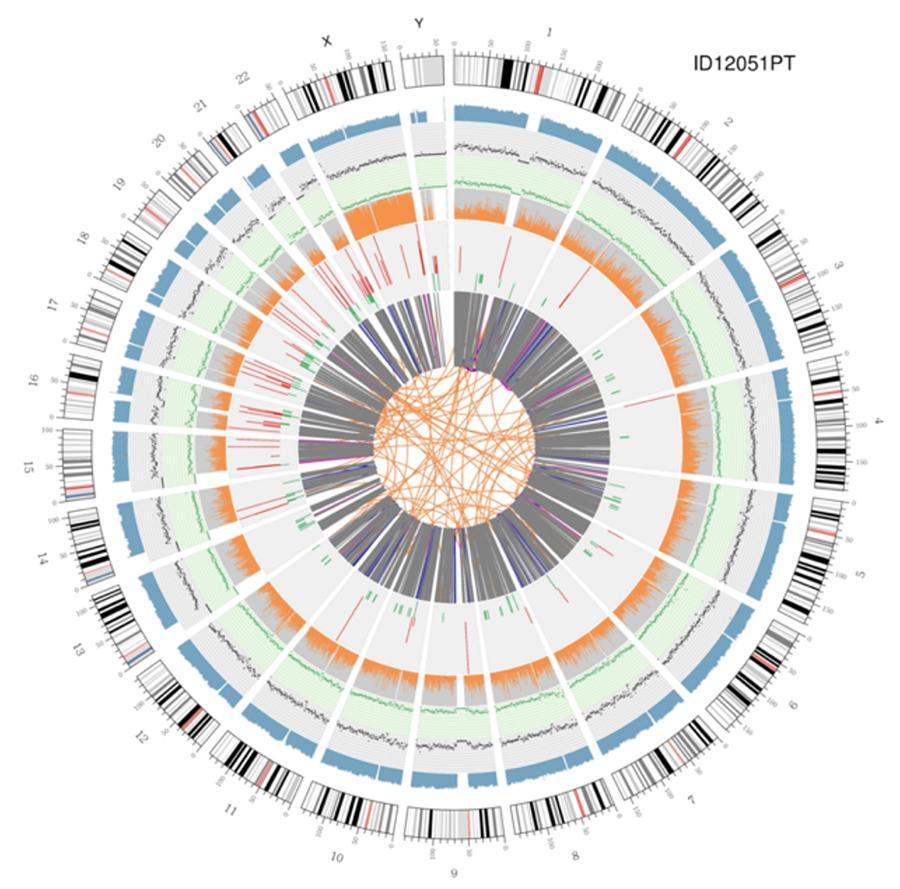
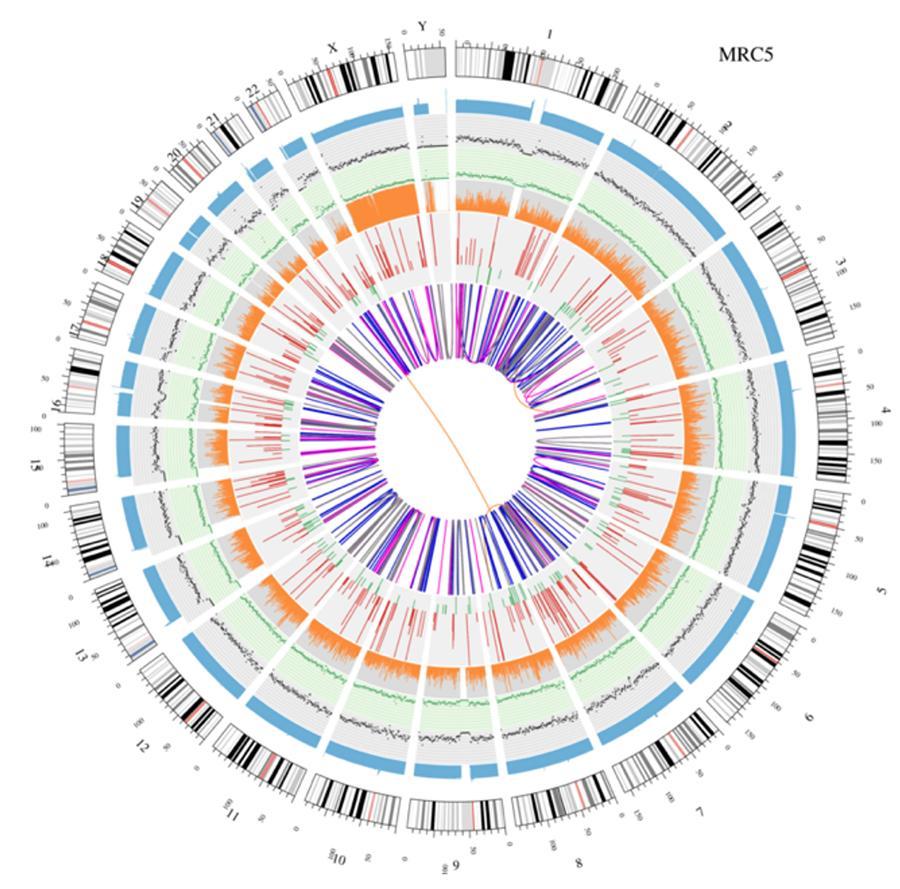
Meaning of the various concentric circles
- The outer circle (first circle) is the chromosome number.
- The second ring (blue) represents the coverage of hystogram-style reads Each hystogram is the average coverage of an area of 0,5 Mbp.
- The third ring (black) represents the density of the INDELs in a 'dispersion graph style'. Each black point is calculated as the number of INDELs, (small insertions/deletions) in a range of 1 Mbp.
- The fourth ring (green) represents the snp density in a 'dispersion graph style'. Each green dot is calculated as the number of SNPs (single nucleotide polymorphisms) in a range of 1 Mbp.
- The fifth ring represents the proportion of homozygous (orange) and heterozygous (grey) SNP in histogram-style. Each histogram is calculated from a 1 Mbp region.
- The sixth ring represents the CNV (copy number variants) inference. Red means DNA pieces gain and green means loss.
- The most central ring represents the SV (structural variants) inference in the exonic and splicing regions. TRA (translocation, orange), INS (insertions, green), DEL (deletions, gray), DUP (duplications, pink), and INV (inversions, blue).
Conclusions
These two genomes show important differences. Specifically, the vaccine genome turns out to be heavily changed compared to that of the cell line deposited in 1966. It is to be said that we did not have the possibility to perform the sequencing of the the cell line used by GlaxoSmithKline for the production of the Priorix Tetra® vaccine, but that of the original stock. Pharmaceutical companies that use cell lines for the production of drugs and vaccines have their own MRC-5 line, but this does not change our suspicions, that is, the fact that genetic stability control is not periodically required is very serious, in our opinion, especially because it is permitted to be present without limit threshold.
Conclusioni
I due genomi mostrano importanti differenze. In particolare, il genoma vaccinale risulta essere pesantemente mutato rispetto a quello della linea cellulare depositata nel 1966. Va detto che non abbiamo avuto la possibilità di sequenziare la linea cellulare che viene utilizzata dalla GlaxoSmithKline per la produzione del vaccino Priorix Tetra, ma quella dello stock originale. Le aziende farmaceutiche che utilizzano linee cellulari per la produzione di farmaci e vaccini, hanno la loro linea MRC-5 ma questo non cambia i nostri sospetti, ovvero il fatto che non venga richiesto periodicamente il controllo della stabilità genetica e questo è gravissimo a nostro avviso, soprattutto perché ne è consentita la presenza senza soglia limite.



